DIOPT Version 9
Website: Version 9
About DIOPT
The identification of orthologs is commonly used for bioinformatics activities such as data mining and establishing models for human diseases. Moreover, our group notes that researchers analyzing the results of screens performed at the Drosophila RNAi Screening Center (DRSC) frequently wish to identify mammalian orthologs of the fly genes that were "hits" (positive results) in their screens.
In helping DRSC screeners to identify orthologs using existing tools and algorithms, we recognized a need for a user-friendly approach to viewing and comparing ortholog predictions obtained using different tools and algorithms. This was our motivation in developing DIOPT. To facilitate identification of orthologs specifically of human disease-associated genes, we further developed DIOPT-DIST. Information about our approaches to development of both tools is summarized below.
The DIOPT Approach
Many tools have emerged to meet the need to identify orthologs. However, low coverage and heterogeneity of these tools present an obstacle to scientists who want to identify a one or a few highest-confidence orthologs for a given gene of interest or conversely, want to cast a wide net and follow up on all possible orthologs of a gene.
Our goal is to provide an easy-to-use resource that facilitates summary, comparison and access to various sources of ortholog predictions. DIOPT integrates human, mouse, fly, worm, zebrafish and yeast ortholog predictions made by Ensembl Compara, HomoloGene, Inparanoid, Isobase, OMA, orthoMCL, Phylome, RoundUp, and TreeFam. DIOPT lets users find ortholog pairs for a specified gene or genes identified by one, many or all of these published approaches. This provides a streamlined method for integration, comparison and access to orthology predictions originating from algorithms based on sequence homology, phylogenetic trees, and functional similarity. DIOPT calculates a simple score indicating the number of tools that support a given orthologous gene-pair relationship, as well as a weighted score based on functional assessment using high quality GO molecular function annotation of all fly-human orthologous pairs predicted by each tool. Differences in the algorithms used by each tool to predict orthologous relationship is one source of difference in the set of predictions made by one tool versus another. However, we also note that some of these differences might be attributable to use of different genome annotation releases used by some tools versus others, and that not all tools cover all of the species that we include in the DIOPT tool (see Tables 1,2 and 3).
DIOPT also displays protein and domain alignments, including percent amino acid identity, for predicted ortholog pairs. These should help you to identify the most appropriate matches among multiple possible orthologs.
The following summary figures and tables help to explain our approach and summarize the tools and algorithms included in DIOPT.
Summary Information and Publications for the Tools Integrated in
Table 1: Summary Information and Publications for the Tools Integrated in DIOPT
| Prediction Method | Source | Prediction Algorithm | Coverage | PMID |
|---|---|---|---|---|
| Compara | Ensembl | Phylogenetic approach | 228 species (vs 104) | 19029536 |
| Homologene | NCBI | Combination of BBH*, tree and synteny | 21 species (vs. 68) | 11125071 |
| Inparanoid | Stockholm University, Sweden | BBH* approach to identify orthologs and in-paralogs | 273 species (vs. 8) | 11743721, 25429972 |
| Isobase | MIT | Sequence and PPI* network alignments | 5 species (vs.2, Nov. 2014) | 21177658 |
| OMA | CBRG, ETH Zurich | BBH*, global sequence alignments | QFO2020 | 17545180 |
| OrthoDB | University of Geneva | Phylogenetic approach | >7302 species (vs.10.1) | 20972218, 25428351 |
| orthoMCL | University of Pennsylvania | Markov Cluster algorithm | 605 species (vs. 6.6) | 12952885 |
| Phylome | Centre for Genomic Regulation (CRG), Spain | Reconstruction of evolutionary histories of all genes in a genome, also known as phylome. | 1059 species,120 Phylomes (vs. 4) QFO2014 | 17962297, 24275491 |
| RoundUp | Harvard Medical School | RSD*, modified BBH* | 2044 species (Apr 2013) | 16777906 |
| TreeFam | Wellcome Trust Sanger Institute | Manually curated based on trees | 109 species (vs. 9) | 16381935, 24194607 |
| Panther | University of Southern California | Phylogenetic approach | 132 species (vs. 16.1) | 26578592 |
| HGNC | European Bioinformatics Institute (EMBL-EBI) | Manually curated | 3 species (Aug 2021) | |
| ZFIN | Zebrafish Model Organism Database | Sequence similarity analysis and manual curation | 4 species (July 2021) | |
| Domainoid | Stockholm University,Sweden | Infer orthology on the domain level | QFO2020 | 31660857 |
| eggNOG | Embl, Germany | Graph-based algorithms | 5090 species (vs. 5.0) | 26582926 |
| OrthoFinder | University of Oxford | Graph-based algorithms | Run code with 12 ref proteome (2020) | 26578592 |
| OrthoInspector | Institut de Génétique et de Biologie Moléculaire et Cellulaire | A novel orthogroup inference algorithm that solves a previously undetected gene length bias in orthogroup inference, resulting in significant improvements in accuracy. | QFO 2020 | 21219603 |
| Hieranoid | Stockholm Bioinformatics Center | The OrthoInspector algorithm is divided into three main steps. First, the results of a Blast all-versus-all (proteomes are blasted against each other) is provided by the user and is parsed to find all the Blast best hits for each protein and to create the groups of inparalogs. Second, the inparalog groups for each organism are compared in a pairwise fashion to define potential orthologs and/or in-paralogs. Third, best hits that contradict the potential orthology between entities are detected. | QFO 2020 | 27742821 |
| ECGA | ECOCYC Genome Database Collection | Manually curated | 2 species (Feb 2021) | NA |
| SGD | Saccharomyces Genome Database | Manually curated | 1 species of paralogs (Oct 2020) | NA |
| sonicParanoid | University of Tokyo | Graph-based algorithms | Run code with 12 ref proteome (2020) | 30032301 |
| NCBI | NIH | Details are not published. Combination of BBH, tree and synteny. | Human2Vertebrate | |
| SwiftOrtho | Iowa State University | Iowa State University | NA | 31648300 |
BBH, Best Blast Hits
RSD, Reciprocal Smallest Distance
PPI, Protein-Protein Interactions
Genome Release Information
Table 2A: Genome Release Information for the Tools Integrated in DIOPT (Version 9)
| Method | Worm | Fish | Fly | Human | Mouse | Yeast | Fission Yeast | Frog | Rat | Thale cress | Mosquito | E coli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compara | WBcel235 | GRCz11 | BDGP6.32 | GRCh38 | GRCm39 | R64-1-1 | NA | Xenopus_tropicalis_v9.1 | Rnor_6.0 | NA | NA | NA |
| Domainoid | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| ECGA | NA | NA | NA | ECGA Feb 2021 | NA | NA | NA | NA | NA | NA | NA | ECGA Feb 2021 |
| eggNOG | WormBase | Ensembl | FlyBase | Ensembl | Ensembl | SGD | PomBase | Ensembl | Ensembl | Araport | VectorBase | Ecocyc |
| HGNC | NA | NA | NA | HGNC Aug 2021 | HGNC Aug 2021 | NA | NA | NA | HGNC Aug 2021 | NA | NA | NA |
| Hieranoid | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| Homologene | WS195 | Zv9 | FlyBase r5.48 | GRCh38 | GRCm38.p2 | R64-1-1 | ASM294v2 | Xtropicalis_v7 | Rnor_5.0 | TAIR10 | AgamP3.3 | NA |
| Inparanoid | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| Isobase | Ensembl v59 | NA | Ensembl v59 | Ensembl v59 | Ensembl v59 | Ensembl v59 | NA | NA | NA | NA | NA | NA |
| OMA | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| orthoDB | Refseq | Refseq | FlyBase | Refseq | Refseq | NA | NA | Refseq | Refseq | Refseq | VectorBase | NA |
| OrthoFinder | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| OrthoInspector | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| orthoMCL | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | HostDB | HostDB | FungiDB | FungiDB | ref proteome 2020 | ref proteome 2020 | ref proteome 2020/td> | VectorBase | ref proteome 2020 |
| Panther | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| Phylome | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| RoundUp | UniProt Apr 2013 | UniProt Apr 2013 | UniProt Apr 2013 | UniProt Apr 2013 | UniProt Apr 2013 | UniProt Apr 2013 | UniProt Apr 2013 | UniProt Apr 2013 | NA | NA | NA | NA |
| SGD | NA | NA | NA | NA | NA | SGD Oct 2020 | NA | NA | NA | NA | NA | NA |
| sonicParanoid | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
| TreeFam | Ensembl v69 | Ensembl v69 | Ensembl v69 | Ensembl v69 | Ensembl v69 | Ensembl v69 | Ensembl v69 | Ensembl v69 | Ensembl v69 | NA | VectorBase | NA |
| ZFIN | NA | ZFIN Aug 2021 | ZFIN Aug 2021 | ZFIN Aug 2021 | ZFIN Aug 2021 | NA | NA | NA | NA | NA | NA | NA |
| NCBI | NA | EntrezGene Oct 2021 | NA | EntrezGene Oct 2021 | EntrezGene Oct 2021 | NA | NA | EntrezGene Oct 2021 | EntrezGene Oct 2021 | NA | NA | NA |
| SwiftOrtho | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 | ref proteome 2020 |
Table 2B: Additional Information About Genome Releases (Version 9)
| Other Resource | Version |
|---|---|
| WormBase | release280 |
| FlyBase | release6.40 |
| RefSeq | release207 (July 2021) |
| EntrezGene | 26-July-2021 |
Table 3. Maximum DIOPT score for each orthologous relationship (Version 9)
| Orthologous Relationship | Max score | Type | Relevant Tools |
|---|---|---|---|
| bacteria-baker's yeast | 12 | ortholog | eggNOG; Panther; orthoMCL; Hieranoid; OMA; Phylome; sonicParanoid; Inparanoid; OrthoFinder; SwiftOrtho; OrthoInspector; Domainoid; |
| bacteria-fish | 12 | ortholog | Panther; SwiftOrtho; Phylome; Domainoid; sonicParanoid; orthoMCL; Hieranoid; OMA; OrthoFinder; eggNOG; OrthoInspector; Inparanoid; |
| bacteria-fission yeast | 12 | ortholog | OrthoInspector; Domainoid; OMA; OrthoFinder; Inparanoid; Hieranoid; SwiftOrtho; eggNOG; orthoMCL; Panther; sonicParanoid; Phylome; |
| bacteria-fly | 12 | ortholog | OMA; sonicParanoid; OrthoInspector; Domainoid; OrthoFinder; Inparanoid; orthoMCL; SwiftOrtho; eggNOG; Hieranoid; Panther; Phylome; |
| bacteria-frog | 11 | ortholog | SwiftOrtho; sonicParanoid; OrthoFinder; Domainoid; OrthoInspector; Hieranoid; Panther; Phylome; OMA; eggNOG; Inparanoid; |
| bacteria-human | 13 | ortholog | orthoMCL; Panther; OrthoFinder; Phylome; Domainoid; Inparanoid; SwiftOrtho; ECGA; OrthoInspector; eggNOG; Hieranoid; sonicParanoid; OMA; |
| bacteria-mosquito | 11 | ortholog | OrthoInspector; Inparanoid; SwiftOrtho; Panther; Domainoid; Phylome; OMA; OrthoFinder; Hieranoid; eggNOG; sonicParanoid; |
| bacteria-mouse | 12 | ortholog | Domainoid; eggNOG; OMA; sonicParanoid; Hieranoid; Panther; Phylome; Inparanoid; SwiftOrtho; orthoMCL; OrthoFinder; OrthoInspector; |
| bacteria-rat | 12 | ortholog | Hieranoid; OrthoInspector; OMA; orthoMCL; Inparanoid; SwiftOrtho; Domainoid; Panther; sonicParanoid; Phylome; eggNOG; OrthoFinder; |
| bacteria-thale cress | 12 | ortholog | OrthoInspector; Hieranoid; Inparanoid; sonicParanoid; Panther; SwiftOrtho; Phylome; eggNOG; OMA; Domainoid; OrthoFinder; orthoMCL; |
| bacteria-worm | 12 | ortholog | OrthoFinder; Panther; Phylome; OrthoInspector; orthoMCL; sonicParanoid; Inparanoid; Domainoid; SwiftOrtho; OMA; eggNOG; Hieranoid; |
| baker's yeast-fish | 16 | ortholog | SwiftOrtho; sonicParanoid; orthoMCL; OMA; Domainoid; Panther; Hieranoid; Phylome; RoundUp; OrthoFinder; TreeFam; eggNOG; OrthoInspector; Homologene; Compara; Inparanoid; |
| baker's yeast-fission yeast | 14 | ortholog | OMA; OrthoInspector; Homologene; Domainoid; Panther; Inparanoid; Phylome; OrthoFinder; orthoMCL; RoundUp; sonicParanoid; TreeFam; eggNOG; Hieranoid; |
| baker's yeast-fly | 16 | ortholog | OrthoInspector; Isobase; orthoMCL; Homologene; Panther; Phylome; Inparanoid; RoundUp; OrthoFinder; Domainoid; TreeFam; OMA; Compara; eggNOG; sonicParanoid; Hieranoid; |
| baker's yeast-frog | 15 | ortholog | SwiftOrtho; OrthoFinder; Panther; Phylome; RoundUp; TreeFam; OrthoInspector; Domainoid; Compara; OMA; sonicParanoid; Homologene; Hieranoid; Inparanoid; eggNOG; |
| baker's yeast-human | 17 | ortholog | Compara; OrthoFinder; Homologene; sonicParanoid; Inparanoid; OMA; SwiftOrtho; Isobase; OrthoInspector; Domainoid; Panther; Phylome; eggNOG; RoundUp; orthoMCL; TreeFam; Hieranoid; |
| baker's yeast-mosquito | 12 | ortholog | Phylome; Homologene; TreeFam; OrthoInspector; Inparanoid; OMA; Domainoid; sonicParanoid; OrthoFinder; eggNOG; Hieranoid; Panther; |
| baker's yeast-mouse | 17 | ortholog | Domainoid; orthoMCL; Isobase; OrthoFinder; Panther; OrthoInspector; Phylome; sonicParanoid; OMA; RoundUp; Hieranoid; TreeFam; Compara; eggNOG; Homologene; Inparanoid; SwiftOrtho; |
| baker's yeast-rat | 15 | ortholog | Hieranoid; OrthoFinder; OMA; eggNOG; Homologene; Panther; Inparanoid; OrthoInspector; orthoMCL; Domainoid; Phylome; SwiftOrtho; TreeFam; sonicParanoid; Compara; |
| baker's yeast-thale cress | 14 | ortholog | Panther; OrthoFinder; sonicParanoid; Domainoid; Phylome; TreeFam; OrthoInspector; Homologene; OMA; Inparanoid; Hieranoid; SwiftOrtho; eggNOG; orthoMCL; |
| baker's yeast-worm | 17 | ortholog | Compara; eggNOG; Hieranoid; OrthoFinder; sonicParanoid; orthoMCL; OrthoInspector; Homologene; Isobase; OMA; Inparanoid; SwiftOrtho; Panther; Phylome; Domainoid; RoundUp; TreeFam; |
| fish-fission yeast | 15 | ortholog | sonicParanoid; Panther; Phylome; RoundUp; OrthoFinder; TreeFam; OMA; orthoMCL; Homologene; Domainoid; Inparanoid; SwiftOrtho; eggNOG; Hieranoid; OrthoInspector; |
| fish-fly | 18 | ortholog | ZFIN; Hieranoid; Panther; OMA; Phylome; sonicParanoid; RoundUp; orthoMCL; TreeFam; OrthoFinder; Compara; Homologene; Inparanoid; SwiftOrtho; Domainoid; eggNOG; OrthoInspector; OrthoDB; |
| fish-frog | 16 | ortholog | OrthoFinder; OMA; Phylome; SwiftOrtho; RoundUp; sonicParanoid; TreeFam; Compara; OrthoInspector; Domainoid; OrthoDB; Homologene; eggNOG; Panther; Hieranoid; Inparanoid; |
| fish-human | 19 | ortholog | Hieranoid; NCBI; ZFIN; OrthoDB; OrthoInspector; Panther; Phylome; Homologene; RoundUp; TreeFam; Inparanoid; Compara; SwiftOrtho; Domainoid; orthoMCL; eggNOG; sonicParanoid; OMA; OrthoFinder; |
| fish-mosquito | 14 | ortholog | sonicParanoid; Homologene; Inparanoid; SwiftOrtho; OrthoDB; OrthoFinder; Panther; Phylome; Domainoid; TreeFam; OMA; eggNOG; OrthoInspector; Hieranoid; |
| fish-mouse | 18 | ortholog | sonicParanoid; ZFIN; Panther; Domainoid; Phylome; orthoMCL; RoundUp; TreeFam; eggNOG; OrthoInspector; Compara; Homologene; Hieranoid; Inparanoid; SwiftOrtho; OMA; OrthoFinder; OrthoDB; |
| fish-rat | 16 | ortholog | Hieranoid; eggNOG; Homologene; OrthoDB; Inparanoid; SwiftOrtho; Panther; sonicParanoid; Phylome; TreeFam; OrthoInspector; orthoMCL; Domainoid; Compara; OrthoFinder; OMA; |
| fish-thale cress | 15 | ortholog | Domainoid; Homologene; Inparanoid; sonicParanoid; OrthoDB; Hieranoid; SwiftOrtho; eggNOG; orthoMCL; Panther; OMA; OrthoFinder; Phylome; TreeFam; OrthoInspector; |
| fish-worm | 17 | ortholog | OMA; orthoMCL; OrthoDB; OrthoInspector; Hieranoid; Panther; Phylome; RoundUp; TreeFam; Homologene; Compara; sonicParanoid; Inparanoid; SwiftOrtho; OrthoFinder; Domainoid; eggNOG; |
| fission yeast-fly | 14 | ortholog | Domainoid; sonicParanoid; Homologene; Hieranoid; Inparanoid; OMA; eggNOG; Panther; Phylome; RoundUp; TreeFam; orthoMCL; OrthoInspector; OrthoFinder; |
| fission yeast-frog | 14 | ortholog | OrthoInspector; OMA; Hieranoid; Panther; Phylome; RoundUp; TreeFam; OrthoFinder; Homologene; Inparanoid; eggNOG; SwiftOrtho; sonicParanoid; Domainoid; |
| fission yeast-human | 15 | ortholog | Domainoid; OMA; Homologene; orthoMCL; Inparanoid; OrthoInspector; Hieranoid; SwiftOrtho; OrthoFinder; eggNOG; sonicParanoid; Panther; Phylome; RoundUp; TreeFam; |
| fission yeast-mosquito | 12 | ortholog | Panther; OMA; OrthoFinder; Phylome; TreeFam; Hieranoid; eggNOG; OrthoInspector; sonicParanoid; Homologene; Inparanoid; Domainoid; |
| fission yeast-mouse | 15 | ortholog | Inparanoid; SwiftOrtho; Hieranoid; orthoMCL; Panther; Phylome; eggNOG; RoundUp; TreeFam; OMA; OrthoInspector; Domainoid; OrthoFinder; Homologene; sonicParanoid; |
| fission yeast-rat | 14 | ortholog | Panther; SwiftOrtho; orthoMCL; Phylome; OrthoInspector; TreeFam; sonicParanoid; Domainoid; eggNOG; Hieranoid; Homologene; OrthoFinder; OMA; Inparanoid; |
| fission yeast-thale cress | 14 | ortholog | Homologene; Panther; orthoMCL; Inparanoid; Phylome; SwiftOrtho; sonicParanoid; TreeFam; OrthoFinder; eggNOG; Domainoid; Hieranoid; OrthoInspector; OMA; |
| fission yeast-worm | 15 | ortholog | OrthoInspector; Domainoid; OMA; Homologene; Inparanoid; sonicParanoid; SwiftOrtho; OrthoFinder; Hieranoid; orthoMCL; eggNOG; Panther; Phylome; RoundUp; TreeFam; |
| fly-frog | 16 | ortholog | OMA; OrthoDB; Hieranoid; Panther; Phylome; RoundUp; OrthoFinder; TreeFam; Compara; sonicParanoid; eggNOG; Homologene; OrthoInspector; Inparanoid; Domainoid; SwiftOrtho; |
| fly-human | 19 | ortholog | Domainoid; Hieranoid; Homologene; OrthoFinder; orthoMCL; sonicParanoid; Inparanoid; OrthoDB; SwiftOrtho; Isobase; eggNOG; Panther; Phylome; RoundUp; TreeFam; OrthoInspector; User_Submission; Compara; OMA; |
| fly-mosquito | 13 | ortholog | Homologene; TreeFam; Inparanoid; Hieranoid; OrthoInspector; eggNOG; sonicParanoid; OrthoDB; OMA; Panther; OrthoFinder; Phylome; Domainoid; |
| fly-mouse | 18 | ortholog | Domainoid; Compara; OrthoInspector; OrthoFinder; Hieranoid; orthoMCL; sonicParanoid; eggNOG; OrthoDB; Isobase; Homologene; Panther; Phylome; Inparanoid; OMA; RoundUp; TreeFam; SwiftOrtho; |
| fly-rat | 16 | ortholog | Hieranoid; OrthoDB; orthoMCL; sonicParanoid; OMA; Panther; Phylome; OrthoFinder; TreeFam; eggNOG; Compara; OrthoInspector; Homologene; Domainoid; Inparanoid; SwiftOrtho; |
| fly-thale cress | 15 | ortholog | Panther; sonicParanoid; Phylome; OrthoFinder; TreeFam; Homologene; Inparanoid; orthoMCL; SwiftOrtho; eggNOG; OrthoInspector; Domainoid; OMA; Hieranoid; OrthoDB; |
| fly-worm | 18 | ortholog | OMA; Domainoid; sonicParanoid; OrthoFinder; Homologene; OrthoDB; Hieranoid; Isobase; Inparanoid; Panther; SwiftOrtho; OrthoInspector; Phylome; RoundUp; eggNOG; TreeFam; orthoMCL; Compara; |
| frog-human | 17 | ortholog | Compara; NCBI; eggNOG; Domainoid; Homologene; Inparanoid; OrthoDB; Hieranoid; SwiftOrtho; OrthoFinder; Panther; OMA; sonicParanoid; Phylome; OrthoInspector; RoundUp; TreeFam; |
| frog-mosquito | 14 | ortholog | Panther; sonicParanoid; Phylome; TreeFam; Homologene; OrthoFinder; Inparanoid; Domainoid; eggNOG; OrthoInspector; SwiftOrtho; Hieranoid; OMA; OrthoDB; |
| frog-mouse | 16 | ortholog | OrthoInspector; Homologene; Domainoid; Inparanoid; SwiftOrtho; Hieranoid; OrthoDB; sonicParanoid; OrthoFinder; Panther; eggNOG; Phylome; RoundUp; TreeFam; OMA; Compara; |
| frog-rat | 15 | ortholog | OMA; Homologene; sonicParanoid; Panther; Inparanoid; Hieranoid; Phylome; SwiftOrtho; TreeFam; OrthoInspector; Compara; eggNOG; OrthoFinder; Domainoid; OrthoDB; |
| frog-thale cress | 14 | ortholog | Panther; eggNOG; Hieranoid; Phylome; TreeFam; Homologene; OrthoInspector; sonicParanoid; Inparanoid; OrthoFinder; SwiftOrtho; OMA; Domainoid; OrthoDB; |
| frog-worm | 16 | ortholog | Compara; eggNOG; Domainoid; OrthoDB; Homologene; sonicParanoid; OrthoInspector; OrthoFinder; Inparanoid; Panther; SwiftOrtho; Phylome; OMA; Hieranoid; RoundUp; TreeFam; |
| human-mosquito | 14 | ortholog | Phylome; TreeFam; Homologene; eggNOG; OMA; Inparanoid; Domainoid; SwiftOrtho; OrthoFinder; OrthoInspector; Hieranoid; OrthoDB; Panther; sonicParanoid; |
| human-mouse | 20 | ortholog | OrthoInspector; OrthoDB; Domainoid; Isobase; OrthoFinder; Panther; Homologene; Phylome; HGNC; RoundUp; Inparanoid; NCBI; TreeFam; Hieranoid; SwiftOrtho; eggNOG; Compara; sonicParanoid; OMA; orthoMCL; |
| human-rat | 18 | ortholog | OMA; Compara; Hieranoid; Homologene; OrthoFinder; sonicParanoid; HGNC; NCBI; Inparanoid; orthoMCL; SwiftOrtho; eggNOG; OrthoInspector; OrthoDB; Panther; Phylome; Domainoid; TreeFam; |
| human-thale cress | 15 | ortholog | Panther; eggNOG; Phylome; TreeFam; OMA; Hieranoid; sonicParanoid; Homologene; Inparanoid; SwiftOrtho; OrthoInspector; OrthoFinder; OrthoDB; Domainoid; orthoMCL; |
| human-worm | 18 | ortholog | OrthoFinder; Hieranoid; Compara; OrthoInspector; orthoMCL; eggNOG; OMA; sonicParanoid; OrthoDB; Isobase; Domainoid; Panther; Homologene; Phylome; Inparanoid; RoundUp; TreeFam; SwiftOrtho; |
| mosquito-mouse | 14 | ortholog | OrthoDB; OrthoFinder; Homologene; Domainoid; Panther; OrthoInspector; Inparanoid; Phylome; SwiftOrtho; TreeFam; eggNOG; OMA; Hieranoid; sonicParanoid; |
| mosquito-rat | 14 | ortholog | OrthoInspector; Panther; OMA; Phylome; TreeFam; Domainoid; Homologene; sonicParanoid; OrthoFinder; Inparanoid; Hieranoid; SwiftOrtho; eggNOG; OrthoDB; |
| mosquito-thale cress | 14 | ortholog | SwiftOrtho; OrthoDB; Panther; sonicParanoid; Phylome; Domainoid; OMA; TreeFam; eggNOG; Hieranoid; Homologene; OrthoInspector; Inparanoid; OrthoFinder; |
| mosquito-worm | 14 | ortholog | eggNOG; Phylome; Homologene; TreeFam; OrthoInspector; Domainoid; OrthoFinder; Inparanoid; SwiftOrtho; Hieranoid; sonicParanoid; OMA; OrthoDB; Panther; |
| mouse-rat | 16 | ortholog | Compara; OMA; OrthoFinder; Homologene; Domainoid; Inparanoid; OrthoInspector; SwiftOrtho; eggNOG; OrthoDB; sonicParanoid; orthoMCL; Hieranoid; Panther; Phylome; TreeFam; |
| mouse-thale cress | 15 | ortholog | eggNOG; OrthoInspector; Inparanoid; Panther; Domainoid; SwiftOrtho; Phylome; OMA; TreeFam; sonicParanoid; Hieranoid; OrthoFinder; orthoMCL; Homologene; OrthoDB; |
| mouse-worm | 18 | ortholog | OrthoFinder; TreeFam; Compara; Homologene; Hieranoid; OrthoInspector; Inparanoid; eggNOG; SwiftOrtho; orthoMCL; OMA; sonicParanoid; OrthoDB; Isobase; Panther; Domainoid; Phylome; RoundUp; |
| rat-thale cress | 15 | ortholog | eggNOG; Phylome; TreeFam; Homologene; OrthoFinder; Inparanoid; OrthoInspector; SwiftOrtho; orthoMCL; sonicParanoid; Domainoid; OMA; OrthoDB; Hieranoid; Panther; |
| rat-worm | 16 | ortholog | OMA; Domainoid; eggNOG; Homologene; OrthoFinder; OrthoDB; OrthoInspector; Inparanoid; SwiftOrtho; Hieranoid; sonicParanoid; Panther; Phylome; TreeFam; Compara; orthoMCL; |
| thale cress-worm | 15 | ortholog | Phylome; orthoMCL; TreeFam; Homologene; sonicParanoid; Domainoid; eggNOG; Inparanoid; SwiftOrtho; OMA; OrthoFinder; OrthoInspector; Hieranoid; OrthoDB; Panther; |
| bacteria-bacteria | 11 | paralog | Inparanoid; SwiftOrtho; Panther; orthoMCL; Phylome; sonicParanoid; Domainoid; OrthoFinder; OMA; OrthoInspector; eggNOG; |
| baker's yeast-baker's yeast | 15 | paralog | Compara; Domainoid; eggNOG; sonicParanoid; Homologene; Isobase; Inparanoid; Panther; Phylome; OrthoInspector; OrthoFinder; RoundUp; OMA; SGD; orthoMCL; |
| fish-fish | 15 | paralog | Inparanoid; OrthoDB; SwiftOrtho; orthoMCL; Panther; Phylome; RoundUp; OrthoFinder; Domainoid; sonicParanoid; Compara; OrthoInspector; OMA; eggNOG; Homologene; |
| fission yeast-fission yeast | 12 | paralog | Homologene; Inparanoid; Panther; eggNOG; Phylome; OMA; RoundUp; OrthoInspector; sonicParanoid; OrthoFinder; Domainoid; orthoMCL; |
| fly-fly | 15 | paralog | Domainoid; OMA; OrthoDB; Isobase; Homologene; Panther; OrthoInspector; Phylome; Inparanoid; RoundUp; eggNOG; Compara; OrthoFinder; sonicParanoid; orthoMCL; |
| frog-frog | 14 | paralog | Inparanoid; sonicParanoid; SwiftOrtho; OrthoDB; Domainoid; Panther; Phylome; eggNOG; RoundUp; Compara; OrthoFinder; OrthoInspector; OMA; Homologene; |
| human-human | 16 | paralog | Compara; OrthoFinder; orthoMCL; OrthoInspector; eggNOG; OMA; Domainoid; OrthoDB; Homologene; Isobase; Inparanoid; Panther; sonicParanoid; SwiftOrtho; Phylome; RoundUp; |
| mosquito-mosquito | 11 | paralog | Panther; Phylome; OMA; Homologene; OrthoFinder; Inparanoid; OrthoInspector; eggNOG; sonicParanoid; Domainoid; OrthoDB; |
| mouse-mouse | 17 | paralog | OrthoInspector; OrthoFinder; sonicParanoid; orthoMCL; OrthoDB; Isobase; Panther; HGNC; Homologene; Phylome; RoundUp; Inparanoid; SwiftOrtho; Compara; Domainoid; eggNOG; OMA; |
| rat-rat | 15 | paralog | orthoMCL; OrthoDB; HGNC; OrthoFinder; Panther; Phylome; eggNOG; Compara; Domainoid; OrthoInspector; OMA; Homologene; sonicParanoid; Inparanoid; SwiftOrtho; |
| thale cress-thale cress | 13 | paralog | Inparanoid; sonicParanoid; SwiftOrtho; OrthoDB; OMA; OrthoInspector; Panther; OrthoFinder; Phylome; Domainoid; orthoMCL; eggNOG; Homologene; |
| worm-worm | 16 | paralog | Homologene; Inparanoid; OrthoInspector; SwiftOrtho; orthoMCL; OrthoDB; Isobase; eggNOG; Domainoid; Panther; sonicParanoid; Phylome; RoundUp; OrthoFinder; Compara; OMA; |
Table 4. DIOPT Weights
DIOPT weights were calculated based on the mean semantic similarity of high quality GO molecular function annotation of all fly-human orthologous pairs predicted by each tool from DIOPT vs1. As such, they do not predict overall performance of an algorithm. Please go to the DIOPT publication for detailed information
(https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-357).| Prediction Method | DIOPT Weight |
|---|---|
| Compara | 0.93 |
| Domainoid | 1 |
| ECGA | 1.5 |
| eggNOG | 0.9 |
| HGNC | 1.5 |
| Hieranoid | 1 |
| Homologene | 1 |
| Inparanoid | 1.05 |
| Isobase | 0.95 |
| OMA | 1.01 |
| OrthoDB | 1.01 |
| OrthoFinder | 1 |
| OrthoInspector | 1 |
| orthoMCL | 0.9 |
| Panther | 1.1 |
| Phylome | 0.91 |
| RoundUp | 1.03 |
| SGD | 1.5 |
| sonicParanoid | 1 |
| TreeFam | 0.96 |
| ZFIN | 1.5 |
| NCBI | 1 |
| SwiftOrtho | 1 |
DIOPT Score Distributions
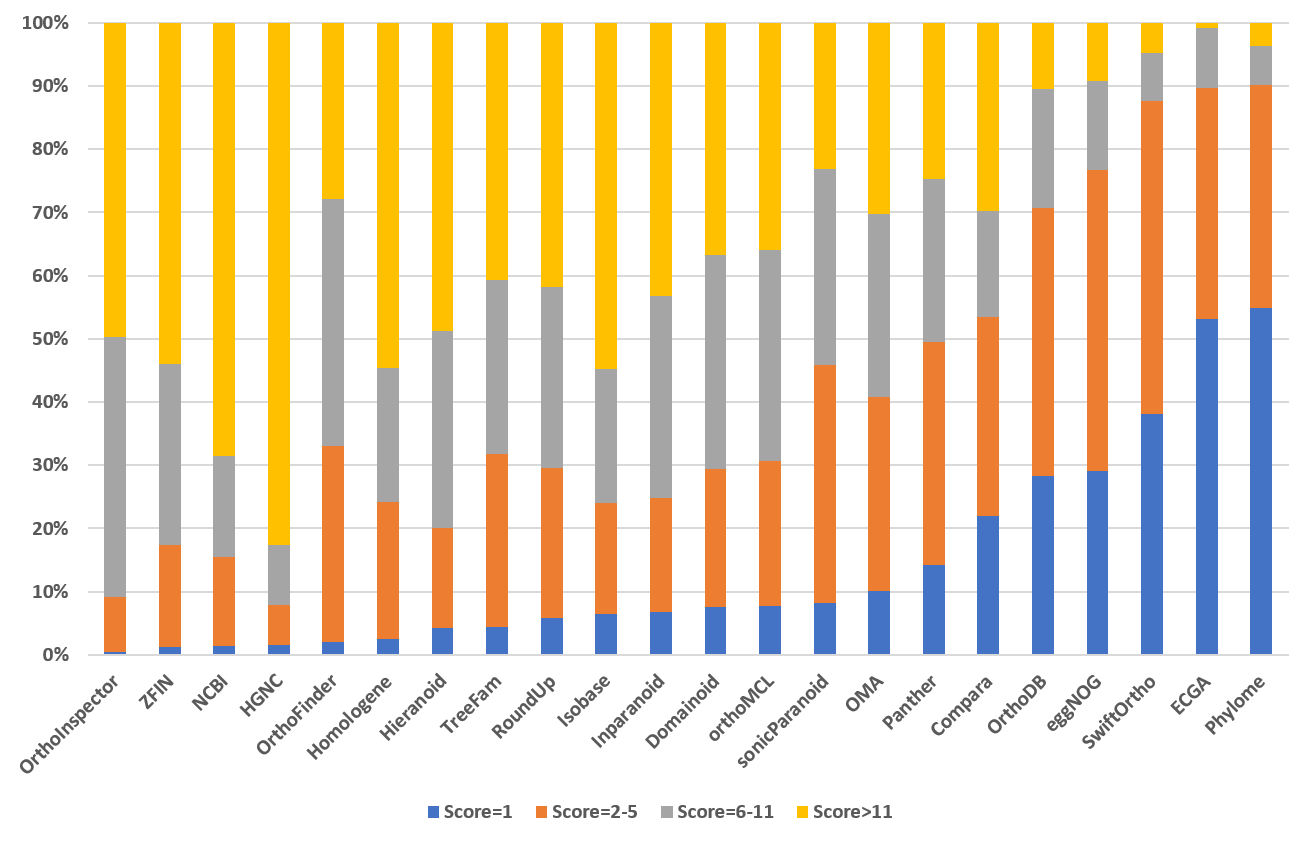
Ortholog Score Distribution (version 9)
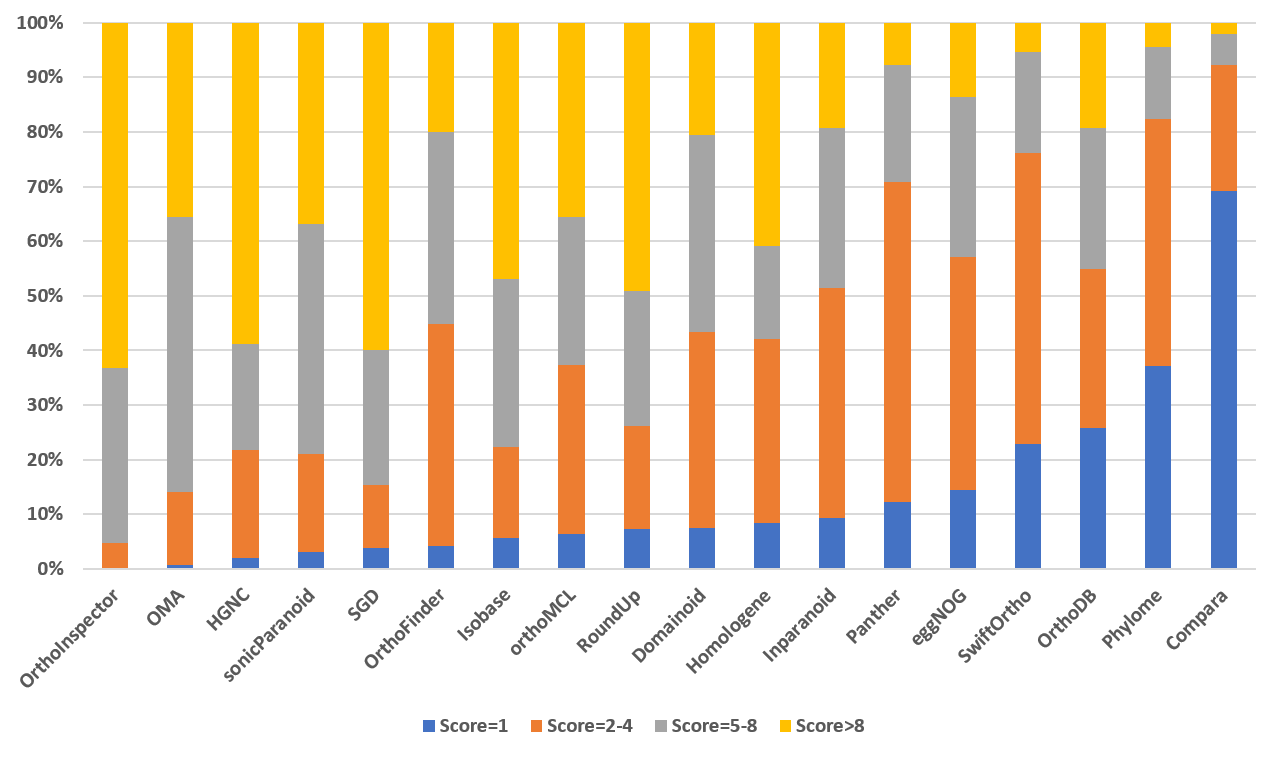
Paralog Score Distribution (version 9)
DIOPT API
You can access diopt data through an API call. For details see:
https://www.flyrnai.org/tools/diopt/web/apiVersions
- Version 10.0 - 2025-
- added tick and removed out-dated predictions
- Version 9.0 - 2023-
- Finalized dataset
- 9.0 (beta) Sept 2021 -
- Revamped UI
- added 2 species: Mosquito (Anopheles gambiae) and E. coli
- added 2 sources of annotation (SGD and ECOCYC) and 2 prediction algorithms (sonicParanoid and Domainid)
- 8.5 August 2021 -
- Revamped UI
- 8.0- August 2019 -
- Updated Data Sources
- 7.1- March 2018 -
- User Feedback on missing relationships
- 7.0- Jan 2018 -
- Updated Data Sources
- added 3 new algorithms: OrthoFinder, OrthoInspector, Hieranoid as a sources
- added new species: Arabidopsis (Thale Cress)
- 6.0.3- Oct 2017 - bug fix to error where in some cases where report fails to generate if certain genes are input
- 6.0.2- June 2017 - fix the ID mapping issue of worm genes to include worm gene related eggNOG predictions
- 6.0.1- Jan 2017 - fixed links to pombebase, column header
- 6.0- Dec 2016 -
- Updated Data Sources
- added eggNOG as a source
- Added paralogs
- 5.5- Oct 2016 - Added multi-sequence alignment from target "All" heatmap
- 5.4- Sept 2016 - Added target species "All" and new filter
- 5.3- May 2016 - Added more prediction tools (Panther, HGNC and ZFIN)
- 5.2.1- April 2016 - Added orthologous rank
- High: best score both ways AND DIOPT score >=2
- Moderate:
- (best score forward or reverse) AND DIOPT score >=2
- DIOPT score >=4
- Low: all others
- 5.2- April 2016 - Added New Species (Rattus norvegicus)
- 5.1.1 - December 2015 - Added Best forward and reverse columns
- 5.1 - November 2015 - Upgraded gene matching algorithm
- 5.0 - November 2015 - Upgraded data sources to version 5