About
The published literature encapsulates years of research about biological systems but finding the relevant literature about a gene or a biological topic is not always straightforward. BioLitMine was implemented to help scientists to quickly mine the literature. Users will able to find MeSH terms as well as the relevant literature. BioLitMine can also help scientists find the principle investigators who are working on a gene of interest or a particular signaling pathway to facilitate the scientific collaborations.
Current features:
- Find associating MeSH terms of given gene, e.g. wg
- Find associating genes of given MeSH term, e.g. "lung cancer", "stem cells"
- Find co-cited genes of input gene
- Find genes list of a given pathway
- Find PIs with publications on a given gene
- Find PIs with publications associated with a given pathway
We are actively working on this tool and more features will be implemented.
How to Use —
Video Tutorial
Instructions
-
Select the type of mapping you are interested in
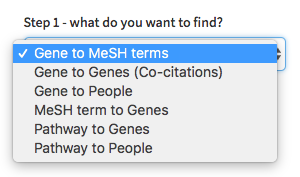
-
Select the model organism of interest
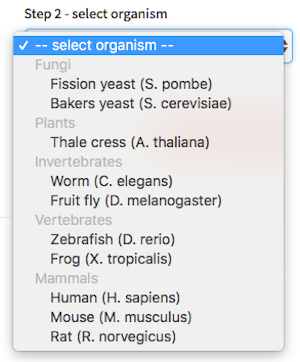
-
Enter your search term -OR- select the pathway of interest
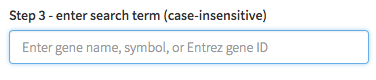
-
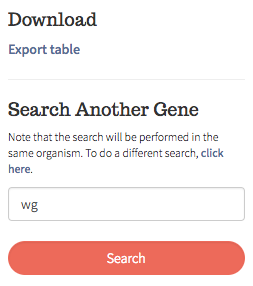
If results are found, you will be re-directed to another page. From there, you can
choose to download your results (as a .csv file) or perform another search
Sources —
Spread the Word —
Cite us
BioLitMine: Advanced Mining of Biomedical and Biological Literature About Human Genes and Genes from Major Model Organisms.
Hu Y, Chung V, Comjean A, Rodiger J, Nipun F, Perrimon N, Mohr SE.G3 (Bethesda). 2020 Oct 7:g3.401775.2020. doi: 10.1534/g3.120.401775. Online ahead of print. PMID: 33028629 (https://pubmed.ncbi.nlm.nih.gov/33028629/)
Our logos
-

244x244 -

244x244