The TRiP found that short hairpin RNAs (shRNAs) containing a 21-bp targeting sequence embedded into a micro-RNA (miR-1) backbone are very effective for gene knockdown in both the germline and soma (Ni et al. 2011). The 2nd generation VALIUM vectors use a microRNA cassette is based on the shRNA design of Haley et al. (2008, 2010) with modifications (Ni et al., 2010). All second generation VALIUM vectors contain vermillion as a selectable marker; an attB sequence to allow phiC31-targeted integration at genomic attP landing sites; two gypsy sequences to enhance hairpin DNA transcription; and two pentamers of UAS, one of which can be excised using the Cre/loxP system to generate a 5XUAS derivative.

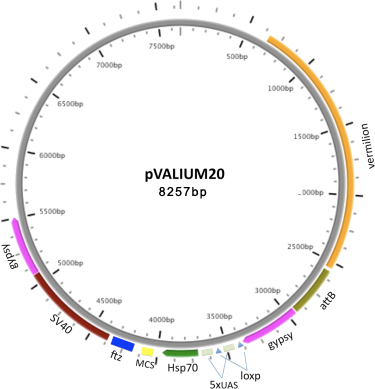
VALIUM20 features the hsp70 basal promoter; a multiple cloning site (MCS) for cloning shRNAs in a miR1 scaffold, and a ftz 3'UTR intron followed by a SV40 3'UTR as a source for a polyA signal sequence. Our data shows that VALIUM20 gives a stronger knockdown than VALIUM10 in the soma, and works well in the germline (Ni et al., 2010).
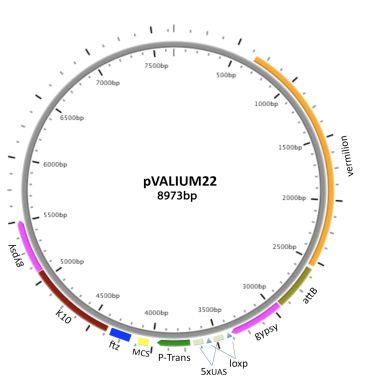
VALIUM22 features the P-transposase core promoter; a multiple cloning site (MCS) for cloning shRNAs in a miR1 scaffold, and a ftz 3'UTR intron followed by a K10 polyA. VALIUM22 triggers effective RNAi knock-down and corresponding phenotypes in the female germline. However, the P-element transposase promoter is less effective than the hsp70 basal promoter for driving expression in somatic cells.
VALIUM21, a variant of VALIUM22, differs only in that it lacks the ftz intron and gypsy sequences found in VALIUM22. It is also effective in the germline, but less so in somatic cells. Only 95 TRiP stocks have been generated in the VALIUM21 vector.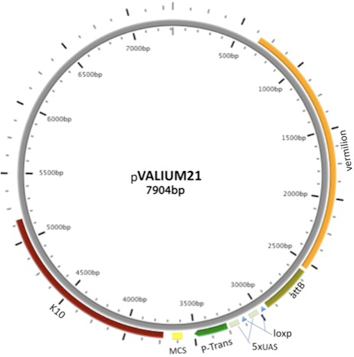
Hairpin Design for 2nd generation VALIUM lines
To design an shRNA for any given gene, the sequences for all exons or portions of exons common to all transcripts are obtained. These sequences are reverse-complemented and all possible 21-bp long sub-sequences are determined. Any sub-sequence with matches to other genes 16 bp-long or longer are removed from consideration. Each sub-sequence has a score calculated based on the formula given by Vert et al. (2006). The top scoring subsequences are selected. A top strand oligo is designed by concatenating "ctagcagt", the sense-strand oligo, "tagttatattcaagcata", the anti-sense oligo, and "gcg". A bottom strand oligo is designed by concatenating "aattcgc", the sense-strand oligo,"tatgcttgaatataacta", the anti-sense oligo, and "actg".
This process is automated by a Perl program developed internally at the TRiP. Primer design can also be done at the DSIR website (http://biodev.extra.cea.fr/DSIR/DSIR.html) using the 21nt siRNA option at default setting. The only difference is that the reverse complement has 2nt offset. We suggest using only the guide strand of DSIR output and then reverse complementing without the 2nt offset.

