Input File Format
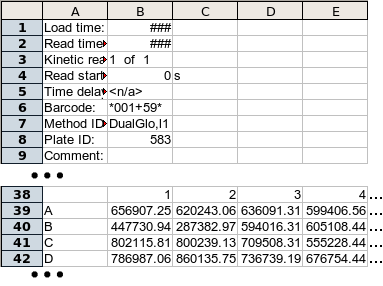
The input file format is based on the output of the Analyst™ Platereader, but the actual format requirements are fairly straightforward and flexible. The file can be a Microsoft Excel™ spreadsheet (.xls) file of a tab-delimitted ASCII text file. The heatmap program will automatically detect which of these file types was used. Note to Open Office users: As of the time this was written, the portion of the program that reads Excel files could not read .xls files created by Open Office Calc, only those created by Microsoft Excel itself. The author of this code, an Open Office user, shares your frustration and is looking for a solution.The file format is a series of plates, each represented by a set of metadata, followed by a matrix of well values, followed by one or more blank rows/lines. The metadata is a series of name-value pairs in columns one and two. The only metadata that is required (or used) is the "Barcode" field, whose name can be any of "Barcode:", "Barcode", "Plate ID:" or "Plate ID". ("Barcode:" has precedence. In the example below, both "Barcode:" and "Plate ID:" are given. The value for "Barcode:" - *001+59* - will be used.) The value associated with the Barcode field is used as a unique name for the plate for display (and for data submission in the internal version.)
The well matrix is a series of rows corresponding to the rows of the plate. The first column contains the row letter and later columns contain well values. The matrix must be preceded by a row with an empty first field and column numbers for the plate in later columns.
Example: