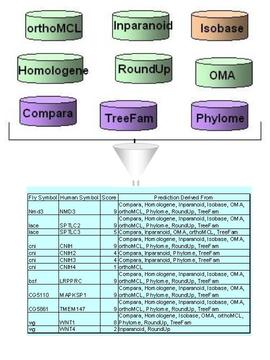
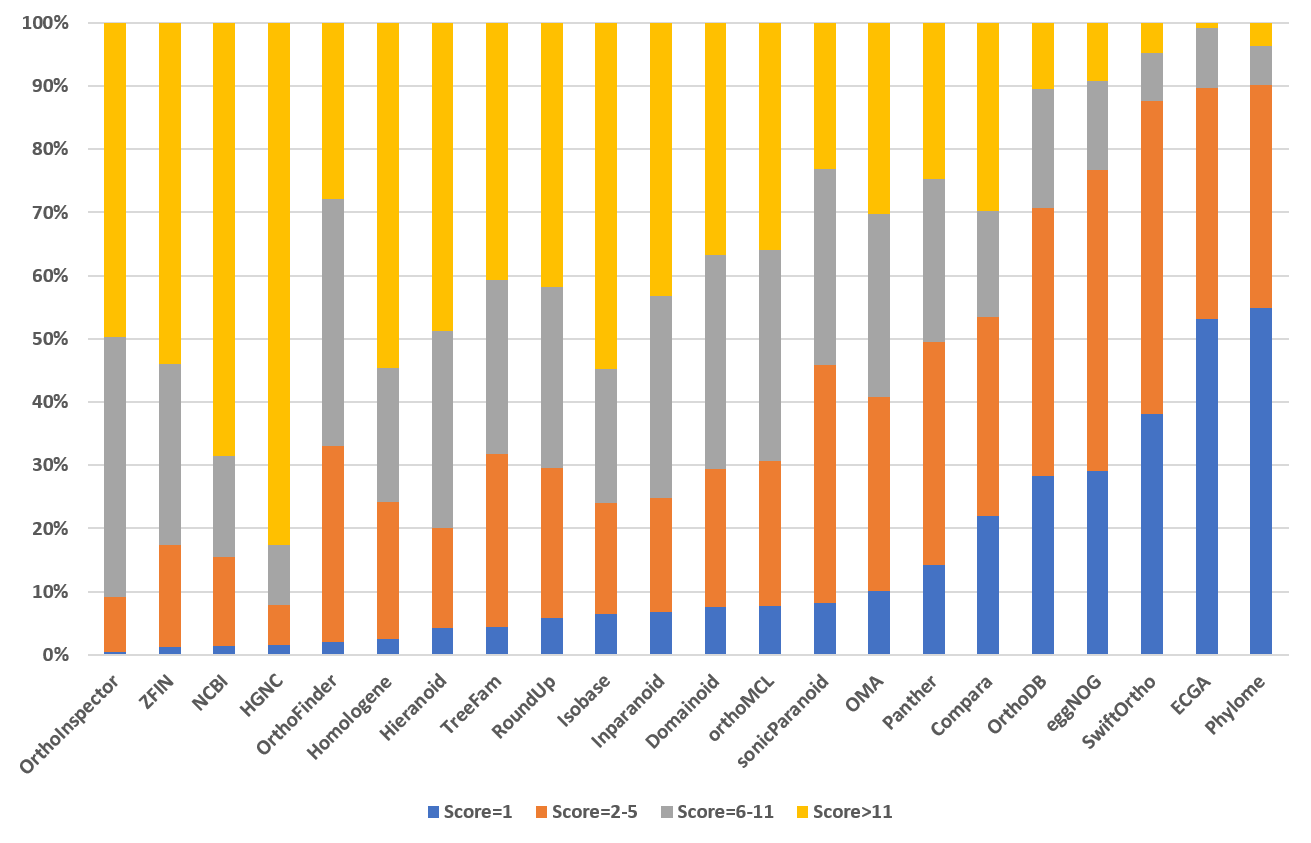
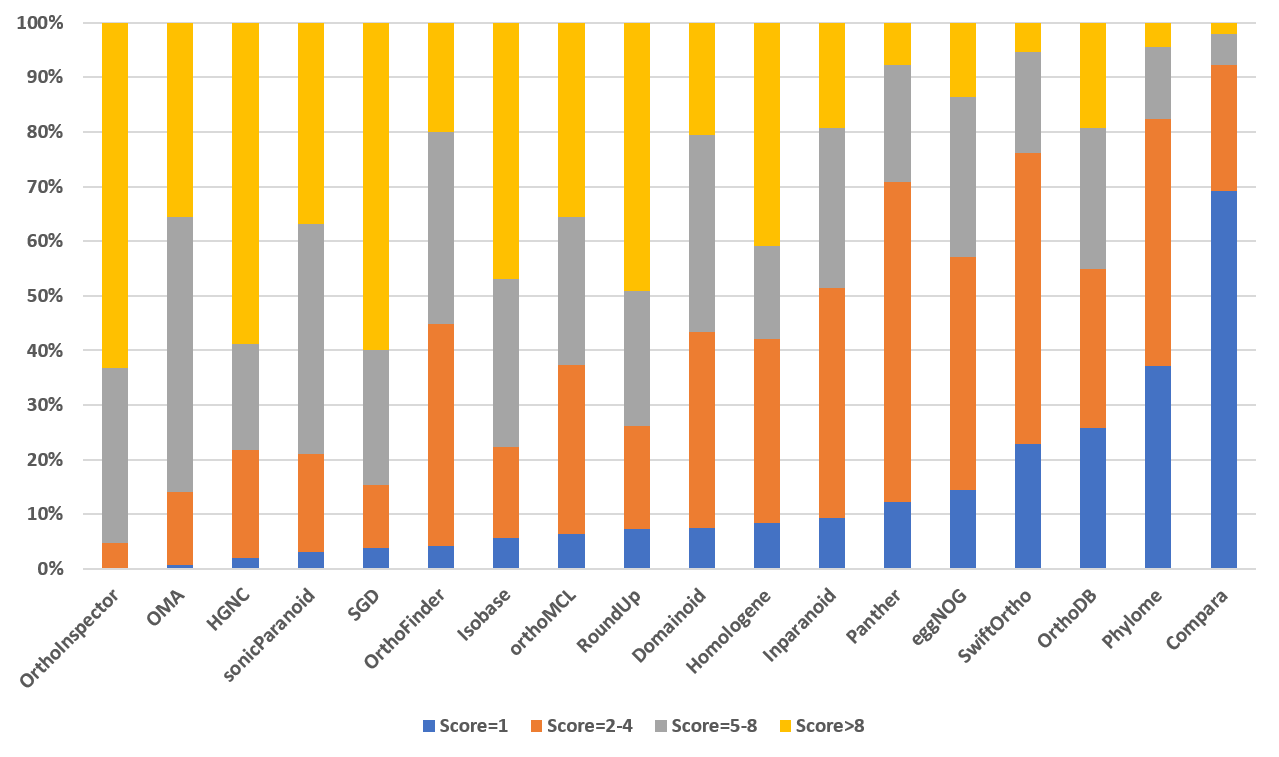
|
Orthologous Relationship
|
Max score
|
Type
|
Relevant Tools
|
|
bacteria-baker's yeast
|
12
|
ortholog
|
eggNOG; Panther; orthoMCL; Hieranoid; OMA; Phylome; sonicParanoid; Inparanoid; OrthoFinder; SwiftOrtho; OrthoInspector; Domainoid;
|
|
bacteria-fish
|
12
|
ortholog
|
Panther; SwiftOrtho; Phylome; Domainoid; sonicParanoid; orthoMCL; Hieranoid; OMA; OrthoFinder; eggNOG; OrthoInspector; Inparanoid;
|
|
bacteria-fission yeast
|
12
|
ortholog
|
OrthoInspector; Domainoid; OMA; OrthoFinder; Inparanoid; Hieranoid; SwiftOrtho; eggNOG; orthoMCL; Panther; sonicParanoid; Phylome;
|
|
bacteria-fly
|
12
|
ortholog
|
OMA; sonicParanoid; OrthoInspector; Domainoid; OrthoFinder; Inparanoid; orthoMCL; SwiftOrtho; eggNOG; Hieranoid; Panther; Phylome;
|
|
bacteria-frog
|
11
|
ortholog
|
SwiftOrtho; sonicParanoid; OrthoFinder; Domainoid; OrthoInspector; Hieranoid; Panther; Phylome; OMA; eggNOG; Inparanoid;
|
|
bacteria-human
|
13
|
ortholog
|
orthoMCL; Panther; OrthoFinder; Phylome; Domainoid; Inparanoid; SwiftOrtho; ECGA; OrthoInspector; eggNOG; Hieranoid; sonicParanoid; OMA;
|
|
bacteria-mosquito
|
11
|
ortholog
|
OrthoInspector; Inparanoid; SwiftOrtho; Panther; Domainoid; Phylome; OMA; OrthoFinder; Hieranoid; eggNOG; sonicParanoid;
|
|
bacteria-mouse
|
12
|
ortholog
|
Domainoid; eggNOG; OMA; sonicParanoid; Hieranoid; Panther; Phylome; Inparanoid; SwiftOrtho; orthoMCL; OrthoFinder; OrthoInspector;
|
|
bacteria-rat
|
12
|
ortholog
|
Hieranoid; OrthoInspector; OMA; orthoMCL; Inparanoid; SwiftOrtho; Domainoid; Panther; sonicParanoid; Phylome; eggNOG; OrthoFinder;
|
|
bacteria-thale cress
|
12
|
ortholog
|
OrthoInspector; Hieranoid; Inparanoid; sonicParanoid; Panther; SwiftOrtho; Phylome; eggNOG; OMA; Domainoid; OrthoFinder; orthoMCL;
|
|
bacteria-worm
|
12
|
ortholog
|
OrthoFinder; Panther; Phylome; OrthoInspector; orthoMCL; sonicParanoid; Inparanoid; Domainoid; SwiftOrtho; OMA; eggNOG; Hieranoid;
|
|
baker's yeast-fish
|
16
|
ortholog
|
SwiftOrtho; sonicParanoid; orthoMCL; OMA; Domainoid; Panther; Hieranoid; Phylome; RoundUp; OrthoFinder; TreeFam; eggNOG; OrthoInspector; Homologene; Compara; Inparanoid;
|
|
baker's yeast-fission yeast
|
14
|
ortholog
|
OMA; OrthoInspector; Homologene; Domainoid; Panther; Inparanoid; Phylome; OrthoFinder; orthoMCL; RoundUp; sonicParanoid; TreeFam; eggNOG; Hieranoid;
|
|
baker's yeast-fly
|
16
|
ortholog
|
OrthoInspector; Isobase; orthoMCL; Homologene; Panther; Phylome; Inparanoid; RoundUp; OrthoFinder; Domainoid; TreeFam; OMA; Compara; eggNOG; sonicParanoid; Hieranoid;
|
|
baker's yeast-frog
|
15
|
ortholog
|
SwiftOrtho; OrthoFinder; Panther; Phylome; RoundUp; TreeFam; OrthoInspector; Domainoid; Compara; OMA; sonicParanoid; Homologene; Hieranoid; Inparanoid; eggNOG;
|
|
baker's yeast-human
|
17
|
ortholog
|
Compara; OrthoFinder; Homologene; sonicParanoid; Inparanoid; OMA; SwiftOrtho; Isobase; OrthoInspector; Domainoid; Panther; Phylome; eggNOG; RoundUp; orthoMCL; TreeFam; Hieranoid;
|
|
baker's yeast-mosquito
|
12
|
ortholog
|
Phylome; Homologene; TreeFam; OrthoInspector; Inparanoid; OMA; Domainoid; sonicParanoid; OrthoFinder; eggNOG; Hieranoid; Panther;
|
|
baker's yeast-mouse
|
17
|
ortholog
|
Domainoid; orthoMCL; Isobase; OrthoFinder; Panther; OrthoInspector; Phylome; sonicParanoid; OMA; RoundUp; Hieranoid; TreeFam; Compara; eggNOG; Homologene; Inparanoid; SwiftOrtho;
|
|
baker's yeast-rat
|
15
|
ortholog
|
Hieranoid; OrthoFinder; OMA; eggNOG; Homologene; Panther; Inparanoid; OrthoInspector; orthoMCL; Domainoid; Phylome; SwiftOrtho; TreeFam; sonicParanoid; Compara;
|
|
baker's yeast-thale cress
|
14
|
ortholog
|
Panther; OrthoFinder; sonicParanoid; Domainoid; Phylome; TreeFam; OrthoInspector; Homologene; OMA; Inparanoid; Hieranoid; SwiftOrtho; eggNOG; orthoMCL;
|
|
baker's yeast-worm
|
17
|
ortholog
|
Compara; eggNOG; Hieranoid; OrthoFinder; sonicParanoid; orthoMCL; OrthoInspector; Homologene; Isobase; OMA; Inparanoid; SwiftOrtho; Panther; Phylome; Domainoid; RoundUp; TreeFam;
|
|
fish-fission yeast
|
15
|
ortholog
|
sonicParanoid; Panther; Phylome; RoundUp; OrthoFinder; TreeFam; OMA; orthoMCL; Homologene; Domainoid; Inparanoid; SwiftOrtho; eggNOG; Hieranoid; OrthoInspector;
|
|
fish-fly
|
18
|
ortholog
|
ZFIN; Hieranoid; Panther; OMA; Phylome; sonicParanoid; RoundUp; orthoMCL; TreeFam; OrthoFinder; Compara; Homologene; Inparanoid; SwiftOrtho; Domainoid; eggNOG; OrthoInspector; OrthoDB;
|
|
fish-frog
|
16
|
ortholog
|
OrthoFinder; OMA; Phylome; SwiftOrtho; RoundUp; sonicParanoid; TreeFam; Compara; OrthoInspector; Domainoid; OrthoDB; Homologene; eggNOG; Panther; Hieranoid; Inparanoid;
|
|
fish-human
|
19
|
ortholog
|
Hieranoid; NCBI; ZFIN; OrthoDB; OrthoInspector; Panther; Phylome; Homologene; RoundUp; TreeFam; Inparanoid; Compara; SwiftOrtho; Domainoid; orthoMCL; eggNOG; sonicParanoid; OMA; OrthoFinder;
|
|
fish-mosquito
|
14
|
ortholog
|
sonicParanoid; Homologene; Inparanoid; SwiftOrtho; OrthoDB; OrthoFinder; Panther; Phylome; Domainoid; TreeFam; OMA; eggNOG; OrthoInspector; Hieranoid;
|
|
fish-mouse
|
18
|
ortholog
|
sonicParanoid; ZFIN; Panther; Domainoid; Phylome; orthoMCL; RoundUp; TreeFam; eggNOG; OrthoInspector; Compara; Homologene; Hieranoid; Inparanoid; SwiftOrtho; OMA; OrthoFinder; OrthoDB;
|
|
fish-rat
|
16
|
ortholog
|
Hieranoid; eggNOG; Homologene; OrthoDB; Inparanoid; SwiftOrtho; Panther; sonicParanoid; Phylome; TreeFam; OrthoInspector; orthoMCL; Domainoid; Compara; OrthoFinder; OMA;
|
|
fish-thale cress
|
15
|
ortholog
|
Domainoid; Homologene; Inparanoid; sonicParanoid; OrthoDB; Hieranoid; SwiftOrtho; eggNOG; orthoMCL; Panther; OMA; OrthoFinder; Phylome; TreeFam; OrthoInspector;
|
|
fish-worm
|
17
|
ortholog
|
OMA; orthoMCL; OrthoDB; OrthoInspector; Hieranoid; Panther; Phylome; RoundUp; TreeFam; Homologene; Compara; sonicParanoid; Inparanoid; SwiftOrtho; OrthoFinder; Domainoid; eggNOG;
|
|
fission yeast-fly
|
14
|
ortholog
|
Domainoid; sonicParanoid; Homologene; Hieranoid; Inparanoid; OMA; eggNOG; Panther; Phylome; RoundUp; TreeFam; orthoMCL; OrthoInspector; OrthoFinder;
|
|
fission yeast-frog
|
14
|
ortholog
|
OrthoInspector; OMA; Hieranoid; Panther; Phylome; RoundUp; TreeFam; OrthoFinder; Homologene; Inparanoid; eggNOG; SwiftOrtho; sonicParanoid; Domainoid;
|
|
fission yeast-human
|
15
|
ortholog
|
Domainoid; OMA; Homologene; orthoMCL; Inparanoid; OrthoInspector; Hieranoid; SwiftOrtho; OrthoFinder; eggNOG; sonicParanoid; Panther; Phylome; RoundUp; TreeFam;
|
|
fission yeast-mosquito
|
12
|
ortholog
|
Panther; OMA; OrthoFinder; Phylome; TreeFam; Hieranoid; eggNOG; OrthoInspector; sonicParanoid; Homologene; Inparanoid; Domainoid;
|
|
fission yeast-mouse
|
15
|
ortholog
|
Inparanoid; SwiftOrtho; Hieranoid; orthoMCL; Panther; Phylome; eggNOG; RoundUp; TreeFam; OMA; OrthoInspector; Domainoid; OrthoFinder; Homologene; sonicParanoid;
|
|
fission yeast-rat
|
14
|
ortholog
|
Panther; SwiftOrtho; orthoMCL; Phylome; OrthoInspector; TreeFam; sonicParanoid; Domainoid; eggNOG; Hieranoid; Homologene; OrthoFinder; OMA; Inparanoid;
|
|
fission yeast-thale cress
|
14
|
ortholog
|
Homologene; Panther; orthoMCL; Inparanoid; Phylome; SwiftOrtho; sonicParanoid; TreeFam; OrthoFinder; eggNOG; Domainoid; Hieranoid; OrthoInspector; OMA;
|
|
fission yeast-worm
|
15
|
ortholog
|
OrthoInspector; Domainoid; OMA; Homologene; Inparanoid; sonicParanoid; SwiftOrtho; OrthoFinder; Hieranoid; orthoMCL; eggNOG; Panther; Phylome; RoundUp; TreeFam;
|
|
fly-frog
|
16
|
ortholog
|
OMA; OrthoDB; Hieranoid; Panther; Phylome; RoundUp; OrthoFinder; TreeFam; Compara; sonicParanoid; eggNOG; Homologene; OrthoInspector; Inparanoid; Domainoid; SwiftOrtho;
|
|
fly-human
|
19
|
ortholog
|
Domainoid; Hieranoid; Homologene; OrthoFinder; orthoMCL; sonicParanoid; Inparanoid; OrthoDB; SwiftOrtho; Isobase; eggNOG; Panther; Phylome; RoundUp; TreeFam; OrthoInspector; User_Submission; Compara; OMA;
|
|
fly-mosquito
|
13
|
ortholog
|
Homologene; TreeFam; Inparanoid; Hieranoid; OrthoInspector; eggNOG; sonicParanoid; OrthoDB; OMA; Panther; OrthoFinder; Phylome; Domainoid;
|
|
fly-mouse
|
18
|
ortholog
|
Domainoid; Compara; OrthoInspector; OrthoFinder; Hieranoid; orthoMCL; sonicParanoid; eggNOG; OrthoDB; Isobase; Homologene; Panther; Phylome; Inparanoid; OMA; RoundUp; TreeFam; SwiftOrtho;
|
|
fly-rat
|
16
|
ortholog
|
Hieranoid; OrthoDB; orthoMCL; sonicParanoid; OMA; Panther; Phylome; OrthoFinder; TreeFam; eggNOG; Compara; OrthoInspector; Homologene; Domainoid; Inparanoid; SwiftOrtho;
|
|
fly-thale cress
|
15
|
ortholog
|
Panther; sonicParanoid; Phylome; OrthoFinder; TreeFam; Homologene; Inparanoid; orthoMCL; SwiftOrtho; eggNOG; OrthoInspector; Domainoid; OMA; Hieranoid; OrthoDB;
|
|
fly-worm
|
18
|
ortholog
|
OMA; Domainoid; sonicParanoid; OrthoFinder; Homologene; OrthoDB; Hieranoid; Isobase; Inparanoid; Panther; SwiftOrtho; OrthoInspector; Phylome; RoundUp; eggNOG; TreeFam; orthoMCL; Compara;
|
|
frog-human
|
17
|
ortholog
|
Compara; NCBI; eggNOG; Domainoid; Homologene; Inparanoid; OrthoDB; Hieranoid; SwiftOrtho; OrthoFinder; Panther; OMA; sonicParanoid; Phylome; OrthoInspector; RoundUp; TreeFam;
|
|
frog-mosquito
|
14
|
ortholog
|
Panther; sonicParanoid; Phylome; TreeFam; Homologene; OrthoFinder; Inparanoid; Domainoid; eggNOG; OrthoInspector; SwiftOrtho; Hieranoid; OMA; OrthoDB;
|
|
frog-mouse
|
16
|
ortholog
|
OrthoInspector; Homologene; Domainoid; Inparanoid; SwiftOrtho; Hieranoid; OrthoDB; sonicParanoid; OrthoFinder; Panther; eggNOG; Phylome; RoundUp; TreeFam; OMA; Compara;
|
|
frog-rat
|
15
|
ortholog
|
OMA; Homologene; sonicParanoid; Panther; Inparanoid; Hieranoid; Phylome; SwiftOrtho; TreeFam; OrthoInspector; Compara; eggNOG; OrthoFinder; Domainoid; OrthoDB;
|
|
frog-thale cress
|
14
|
ortholog
|
Panther; eggNOG; Hieranoid; Phylome; TreeFam; Homologene; OrthoInspector; sonicParanoid; Inparanoid; OrthoFinder; SwiftOrtho; OMA; Domainoid; OrthoDB;
|
|
frog-worm
|
16
|
ortholog
|
Compara; eggNOG; Domainoid; OrthoDB; Homologene; sonicParanoid; OrthoInspector; OrthoFinder; Inparanoid; Panther; SwiftOrtho; Phylome; OMA; Hieranoid; RoundUp; TreeFam;
|
|
human-mosquito
|
14
|
ortholog
|
Phylome; TreeFam; Homologene; eggNOG; OMA; Inparanoid; Domainoid; SwiftOrtho; OrthoFinder; OrthoInspector; Hieranoid; OrthoDB; Panther; sonicParanoid;
|
|
human-mouse
|
20
|
ortholog
|
OrthoInspector; OrthoDB; Domainoid; Isobase; OrthoFinder; Panther; Homologene; Phylome; HGNC; RoundUp; Inparanoid; NCBI; TreeFam; Hieranoid; SwiftOrtho; eggNOG; Compara; sonicParanoid; OMA; orthoMCL;
|
|
human-rat
|
18
|
ortholog
|
OMA; Compara; Hieranoid; Homologene; OrthoFinder; sonicParanoid; HGNC; NCBI; Inparanoid; orthoMCL; SwiftOrtho; eggNOG; OrthoInspector; OrthoDB; Panther; Phylome; Domainoid; TreeFam;
|
|
human-thale cress
|
15
|
ortholog
|
Panther; eggNOG; Phylome; TreeFam; OMA; Hieranoid; sonicParanoid; Homologene; Inparanoid; SwiftOrtho; OrthoInspector; OrthoFinder; OrthoDB; Domainoid; orthoMCL;
|
|
human-worm
|
18
|
ortholog
|
OrthoFinder; Hieranoid; Compara; OrthoInspector; orthoMCL; eggNOG; OMA; sonicParanoid; OrthoDB; Isobase; Domainoid; Panther; Homologene; Phylome; Inparanoid; RoundUp; TreeFam; SwiftOrtho;
|
|
mosquito-mouse
|
14
|
ortholog
|
OrthoDB; OrthoFinder; Homologene; Domainoid; Panther; OrthoInspector; Inparanoid; Phylome; SwiftOrtho; TreeFam; eggNOG; OMA; Hieranoid; sonicParanoid;
|
|
mosquito-rat
|
14
|
ortholog
|
OrthoInspector; Panther; OMA; Phylome; TreeFam; Domainoid; Homologene; sonicParanoid; OrthoFinder; Inparanoid; Hieranoid; SwiftOrtho; eggNOG; OrthoDB;
|
|
mosquito-thale cress
|
14
|
ortholog
|
SwiftOrtho; OrthoDB; Panther; sonicParanoid; Phylome; Domainoid; OMA; TreeFam; eggNOG; Hieranoid; Homologene; OrthoInspector; Inparanoid; OrthoFinder;
|
|
mosquito-worm
|
14
|
ortholog
|
eggNOG; Phylome; Homologene; TreeFam; OrthoInspector; Domainoid; OrthoFinder; Inparanoid; SwiftOrtho; Hieranoid; sonicParanoid; OMA; OrthoDB; Panther;
|
|
mouse-rat
|
16
|
ortholog
|
Compara; OMA; OrthoFinder; Homologene; Domainoid; Inparanoid; OrthoInspector; SwiftOrtho; eggNOG; OrthoDB; sonicParanoid; orthoMCL; Hieranoid; Panther; Phylome; TreeFam;
|
|
mouse-thale cress
|
15
|
ortholog
|
eggNOG; OrthoInspector; Inparanoid; Panther; Domainoid; SwiftOrtho; Phylome; OMA; TreeFam; sonicParanoid; Hieranoid; OrthoFinder; orthoMCL; Homologene; OrthoDB;
|
|
mouse-worm
|
18
|
ortholog
|
OrthoFinder; TreeFam; Compara; Homologene; Hieranoid; OrthoInspector; Inparanoid; eggNOG; SwiftOrtho; orthoMCL; OMA; sonicParanoid; OrthoDB; Isobase; Panther; Domainoid; Phylome; RoundUp;
|
|
rat-thale cress
|
15
|
ortholog
|
eggNOG; Phylome; TreeFam; Homologene; OrthoFinder; Inparanoid; OrthoInspector; SwiftOrtho; orthoMCL; sonicParanoid; Domainoid; OMA; OrthoDB; Hieranoid; Panther;
|
|
rat-worm
|
16
|
ortholog
|
OMA; Domainoid; eggNOG; Homologene; OrthoFinder; OrthoDB; OrthoInspector; Inparanoid; SwiftOrtho; Hieranoid; sonicParanoid; Panther; Phylome; TreeFam; Compara; orthoMCL;
|
|
thale cress-worm
|
15
|
ortholog
|
Phylome; orthoMCL; TreeFam; Homologene; sonicParanoid; Domainoid; eggNOG; Inparanoid; SwiftOrtho; OMA; OrthoFinder; OrthoInspector; Hieranoid; OrthoDB; Panther;
|
|
bacteria-bacteria
|
11
|
paralog
|
Inparanoid; SwiftOrtho; Panther; orthoMCL; Phylome; sonicParanoid; Domainoid; OrthoFinder; OMA; OrthoInspector; eggNOG;
|
|
baker's yeast-baker's yeast
|
15
|
paralog
|
Compara; Domainoid; eggNOG; sonicParanoid; Homologene; Isobase; Inparanoid; Panther; Phylome; OrthoInspector; OrthoFinder; RoundUp; OMA; SGD; orthoMCL;
|
|
fish-fish
|
15
|
paralog
|
Inparanoid; OrthoDB; SwiftOrtho; orthoMCL; Panther; Phylome; RoundUp; OrthoFinder; Domainoid; sonicParanoid; Compara; OrthoInspector; OMA; eggNOG; Homologene;
|
|
fission yeast-fission yeast
|
12
|
paralog
|
Homologene; Inparanoid; Panther; eggNOG; Phylome; OMA; RoundUp; OrthoInspector; sonicParanoid; OrthoFinder; Domainoid; orthoMCL;
|
|
fly-fly
|
15
|
paralog
|
Domainoid; OMA; OrthoDB; Isobase; Homologene; Panther; OrthoInspector; Phylome; Inparanoid; RoundUp; eggNOG; Compara; OrthoFinder; sonicParanoid; orthoMCL;
|
|
frog-frog
|
14
|
paralog
|
Inparanoid; sonicParanoid; SwiftOrtho; OrthoDB; Domainoid; Panther; Phylome; eggNOG; RoundUp; Compara; OrthoFinder; OrthoInspector; OMA; Homologene;
|
|
human-human
|
16
|
paralog
|
Compara; OrthoFinder; orthoMCL; OrthoInspector; eggNOG; OMA; Domainoid; OrthoDB; Homologene; Isobase; Inparanoid; Panther; sonicParanoid; SwiftOrtho; Phylome; RoundUp;
|
|
mosquito-mosquito
|
11
|
paralog
|
Panther; Phylome; OMA; Homologene; OrthoFinder; Inparanoid; OrthoInspector; eggNOG; sonicParanoid; Domainoid; OrthoDB;
|
|
mouse-mouse
|
17
|
paralog
|
OrthoInspector; OrthoFinder; sonicParanoid; orthoMCL; OrthoDB; Isobase; Panther; HGNC; Homologene; Phylome; RoundUp; Inparanoid; SwiftOrtho; Compara; Domainoid; eggNOG; OMA;
|
|
rat-rat
|
15
|
paralog
|
orthoMCL; OrthoDB; HGNC; OrthoFinder; Panther; Phylome; eggNOG; Compara; Domainoid; OrthoInspector; OMA; Homologene; sonicParanoid; Inparanoid; SwiftOrtho;
|
|
thale cress-thale cress
|
13
|
paralog
|
Inparanoid; sonicParanoid; SwiftOrtho; OrthoDB; OMA; OrthoInspector; Panther; OrthoFinder; Phylome; Domainoid; orthoMCL; eggNOG; Homologene;
|
|
worm-worm
|
16
|
paralog
|
Homologene; Inparanoid; OrthoInspector; SwiftOrtho; orthoMCL; OrthoDB; Isobase; eggNOG; Domainoid; Panther; sonicParanoid; Phylome; RoundUp; OrthoFinder; Compara; OMA;
|